Estimating Distances Between Genes
Estimating Linkage From Three-Point Crosses
Recombination Involves Exchange Of Chromosomal Material
Lod Score Method
Lod Score Method of Estimating Linkage Distances
The following pedigree will be used to demonstrate another method developed to determine the distance between genes. This approach has been widely adapted to various system and genetic programs have been developed based on this technique. First let's look at the pedigree below.
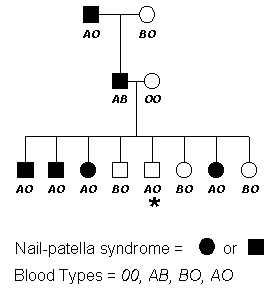
Several points should be drawn from this pedigree. Even though we are working with the same two genes, nail-patella and blood type, in this pedigree the dominant allele seems to be coupled with the A blood type allele. Remember in the previous example, the dominant nail-patella allele was linked with the B allele. This is an important point in genetics --- not all linkages between alleles of two genes are found to be constant throughout a species. Why??? Because at some point in the lineage of this family, the disease (nail-patella) allele recombined and became linked to a different blood type allele. In even other lineages, the nail-patella causing allele is linked to the O blood type allele.
Next let's determine the linkage distance between the two genes. As you can see, we have one recombinant among the eight progeny. This gives us a recombination frequency of 0.125 and a distance of 12.5 cM.
We will now introduce a new method to calculate linkage distances called the Lod Score Method. The method developed by Newton E. Morton is an iterative approach were a series of lod scores are calculated from a number of proposed linkage distance. Here is how the method works. A linkage distance is estimated, and given that estimate, the probablity of a given birth sequence is calculated. That value is then divided by the probability of a given birth sequence assuming that the genes are unlinked. The log of this value is calculated, and that value is the lod score for this linkage distance estimate. The same process is repeated with another linkage distance estimate. A series of these lod scores are obtained using different linkage distances, and the linkage distance giving the highest Lod score is considered the estimate of the linkage distance. The following is the formula for the lod score:

The above example will be used to demonstrate the principle. We will first use 0.125 as our estimate of the recombination fraction. In this first birth sequence, we have an individual with a parental genotype.The probablity of this event is (1 - 0.125). Because there are two parental types, this value is divided by two to give a value of 0.4375. In this pedigree we have a total of seven parental types. We also have one recombinant type. The probability of this event is 0.125 which is divided by two because two recombinant types exist.
What would the sequence of births be if these genes were unlinked? When two genes are unlinked the recombination frequency is 0.5. Therefore, the probability of any given genotype would be 0.25.
Now let's put the whole method together. The probability of a given birth sequence is the product of each of the independent events.So the probability of the birth sequence based on our estimate of 0.125 as the recombination frequence would be equal to (0.4375)7(0.0625)1 = 0.0001917. The probability of the birth sequence based on no linkage would be (0.25)8 = 0.0000153. Now divide the linkage probability by the non-linkage probability and you get a value of 12.566. Next take the log of this value, and you obtain a value of 1.099. This value is the lod score.
As was mentioned, this is repeated for a series of recombination frequency estimates. The table below gives the lod score for six different linkage estimates.
|
Recombination Frequency |
Lod Score |
|---|---|
|
0.050 |
0.951 |
|
0.100 |
1.088 |
|
0.125 |
1.099 |
|
0.150 |
1.090 |
|
0.200 |
1.031 |
|
0.250 |
0.932 |
As the table shows, the largest lod score corresponds to a linkage estimate of 0.125. In practice, we would like to see a lod score greater that 3.0. What this means is that the likelihood of linkage occurring at this distance is 1000 times greater that no linkage.
The lod score is a widely used technique not only in human research but in plant and animal linkage analyzes as well. An important software package, MAPMAKER, that is widely used in plant mapping research is based in part on the lod score method.
Copyright © 1998. Phillip McClean