Dynamic Prairies:
Collaboration to understand one of the most threatened ecosystems in America
The National Science Foundation's National Ecological Observatory Network (NEON) is a continental-scale observation facility operated by Battelle and designed to collect long-term open access ecological data to better understand how U.S. ecosystems are changing. Northern Prairie Wildlife Research Center is one of seventeen USGS Science Centers that develop and disseminate the scientific information needed to understand, conserve, and manage the Nation’s rich biological resources. An intern with the RaMP project could participate in data collection at our field sites and learn to utilize long-term data to explore questions in a wide variety of ecological areas.
Potential research questions:
- Cattle vs. mice-How do land management practices impact small mammal populations?
- Population rollercoaster- What causes the booms and busts in fish populations?
- Source or sink- how can prairies help fight climate change?
Field and lab work: The type of fieldwork will depend on your experience and interest in terrestrial (i.e. fauna and flora) or aquatic ecology. Fieldwork will include learning a wide variety of protocols for sampling and data collection. Lab experiences can include identifying and processing plant, soil and invertebrate samples, measuring pH and conducting titrations. Depending on the research question there might also be an opportunity to develop conduct a field experiment to supplement the long-term datasets.
Instrumentation: Interns have the opportunity to help maintain field site instrumentation, which includes calibrating sensors, troubleshooting problems, site maintenance, and assisting with corrective maintenance of sensors.
Skill development: Increase your understanding of ecological processes and change at large spatial and temporal scales. In addition to hands on experience in a wide variety of ecological data collection, we offer educational resources to gain the data skills (R coding skills) needed to work with NEON data. You would have the opportunity to develop a project using NEON data and potentially make connections to data collected at other sites around the country to answer important ecological questions.
Network Mentors: Andrea Anteau, NEON and Kyle McLean, USGS
Insults for free: The role of metamorphosis in reversing aging in bees
Insects that go through metamorphosis are an extreme example of disposable somatic tissue, because juvenile tissues are destroyed during metamorphosis, while the adult body is built from imaginal, stem cell-like tissues. Metamorphosis may allow insects to take “insults for free” during the larval stage—if tissue accumulates stress-related damage, the individual only has to make it to metamorphosis, then the tissue is disposed of and no longer influences fitness. We hypothesize that metamorphosis allows insects to compartmentalize aging, only investing in regenerative maintenance of tissues that are not disposed of during metamorphosis, while allowing rapid aging of disposable somatic tissues. In addition, we will test to what extent stress affects the rate of aging in these tissues. We will test this hypothesis by comparing cellular aging in brain (not disposable), hemolymph (constantly regenerated), reproductive organs (not soma, not disposable), epidermis and larval muscle (disposable), and flight muscle (regenerated anew). Our preliminary data show that telomeres in prepupae are shorter than those of adults. However, that data compares whole-body prepupae to adult flight muscle, a tissue that could be governed by different aging dynamics than other adult tissues.
We will test the ability of metamorphosis to reset the aging effects of stress by treating prepupal M. rotundata with chemicals to increase and decrease oxidative stress. We will sample tissues in prepupae 24 hours after treatment and when the bees emerge as adults. We will sample brain, hemolymph, ovary/teste, and epidermis. In the adults, we will also sample flight muscle. For all of these tissues, we will measure telomere length, DNA damage, lipid peroxidation, glutathione activity, and reactive oxygen species. Adult bees will also be tested for performance upon emergence and rate of senescence by measuring fight performance by a drop test, non-feeding lifespan, and activity during the second week post-emergence.
Experimental tools and skills/methods: Insect husbandry, Programming incubators, Molecular biology methods including: qPCR and biochemical assays, Organization of experimental samples and data, 3D printing and designing equipment for the experiment, Dissection and microscopy
Network Mentors: Julia Bowsher, NDSU, Kendra Greenlee, NDSU, and Britt Heidinger, NDSU
Genetic versus phenotypic divergence
The RaMP member will lead a project examining how behavior and morphology are genetically linked. By collecting data on the morphology of crickets—body size and shape—and combining this with behavioral data, the RaMP member will determine the evolutionary potential of natural populations to respond to changing environments.
The RaMP member will lead a project examining how populations diverge genetically versus in morphology and behavior. By combining data on the phenotypes of multiple populations of crickets with the extraction and analysis of genetic information from preserved specimens, the RaMP member will quantify how evolution has shaped both what individuals do and underlying genetic variation.
Patterns of correlations of among traits like behaviors and morphology can constrain the evolution of populations and maintain phenotypic similarity of genetically isolated populations. How this then maps to underlying genetic divergence is not necessarily clear and can also identify genomic regions important for phenotypes.
The RaMP member will learn methods for isolating DNA from organisms, methods of analysis for genomic data, and methods for comparing phenotypic and genetic variation among populations.
As part of this project, we will train the RaMP member in data collection, DNA extraction, genomic analyses, project and data management, statistical analyses, and data science. We will also train the RaMP member in the curation of large datasets, how to properly plan statistical analyses given research questions and data structure, how to code analytical pipelines (using the R statistical language), and how to properly interpret outcomes of statistical analyses. Through presentations of results in local and professional meetings, the member will also gain experience in presenting scientific results verbally, textually, and graphically.
Network Mentor: Ned Dochtermann, North Dakota State University
Elucidating the genetics of rhizobium-legume symbiosis to enable nitrogen-fixing cereal crops
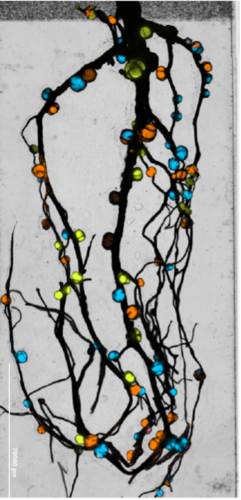
Legume crops like peas or soybeans are important for sustainable agriculture because of their ability to form a symbiotic relationship with bacteria called rhizobia. Rhizobia elicit the formation of new organs on plant roots called root nodules (engineered to fluoresce in the photo) wherein they transform nitrogen from the air into useable forms for the plant, replacing the need for nitrogen fertilization in agriculture. Unfortunately this symbiosis has only evolved in legume crops and is currently not accessibly by grain crops critical to the global food system such as wheat, corn and rice. A “holy grail” of synthetic biology is to transfer the nitrogen-fixing associations between rhizobia and legumes to cereal crops. To do this we must gain a detailed understanding of the genetic programs in rhizobia and legumes required to carry out an efficient symbiosis. Students will be involved in research projects to decipher the genes involved in effective rhizobium symbioses, and engineering new-to-nature rhizobium symbioses, with a long-term goal of engineering nitrogen-fixation in cereal crops. This work is intensely multidisciplinary and students will gain experience in diverse approaches such as next-generation sequencing of DNA, molecular cloning, microbial engineering, fluorescent microscopy, and bioinformatics. Legume crops like peas or soybeans are important for sustainable agriculture because of their ability to form a symbiotic relationship with bacteria called rhizobia. Rhizobia elicit the formation of new organs on plant roots called root nodules (engineered to fluoresce in the photo) wherein they transform nitrogen from the air into useable forms for the plant, replacing the need for nitrogen fertilization in agriculture. Unfortunately this symbiosis has only evolved in legume crops and is currently not accessibly by grain crops critical to the global food system such as wheat, corn and rice. A “holy grail” of synthetic biology is to transfer the nitrogen-fixing associations between rhizobia and legumes to cereal crops. To do this we must gain a detailed understanding of the genetic programs in rhizobia and legumes required to carry out an efficient symbiosis. Students will be involved in research projects to decipher the genes involved in effective rhizobium symbioses, and engineering new-to-nature rhizobium symbioses, with a long-term goal of engineering nitrogen-fixation in cereal crops. This work is intensely multidisciplinary and students will gain experience in diverse approaches such as next-generation sequencing of DNA, molecular cloning, microbial engineering, fluorescent microscopy, and bioinformatics.
Network Mentors: Barney Geddes, NDSU
Ecology and Genetics of a Co-Managed Elk Herd on Tribal Lands
United Tribes Technical College (UTTC) is investigating a co-managed elk herd with North Dakota Game & Fish Department and Standing Rock Game & Fish Department. The herd has expanded in the past ten years from about a dozen animals to well over 200 individuals. The long-term overarching project involves identifying seasonal differences in elk diet and gut microbiome, and statewide population genetics that compare to current genetic projects in Minnesota and Montana. We do not have a solid understanding of the origin of the Standing Rock elk herd and there appears to be indications of a bottleneck effect resulting in antler deformities in a percentage of the population. This contributes to impacts on cultural uses of elk antlers, the Tribe’s potential for ecotourism, and the Tribe and State’s development of trophy hunting which can sustain conservation efforts.
The CHANGE RaMP research fellow project will focus on:
- Developing a protocol for annual analysis of elk jaw samples from ND Game & Fish Department. The department annually collects jaw samples from across the state that are sent to labs for aging. They are sharing duplicates of the samples with UTTC so that we can obtain additional genetic/genomic data.
- Conducting a pilot study of analyzing (a) dietary components in elk pellets that can be replicated during different seasons and (b) examining elk DNA from intestinal mucus coating the pellets.
This project would be a good mix of field experience and lab work. Field experiences will include establishing transects, identifying plants and animal species, and collecting pellets. Some of the locations will be remote and off-trail in extreme summer conditions in mixed-grass prairies and badlands ecosystems. Lab experiences will include DNA Extraction, PCR, miSeq sequencing, and bioinformatics.
Network Mentors: Jeremy Guinn, United Tribes Technical College
Bats on the Brink
This research initiative focuses on addressing the decline in bat populations in the United States, specifically examining the impact of White-Nose Syndrome (WNS) on endangered bat species, including the Northern Long-Eared Bat (NLEB). With these species federally listed as endangered, monitoring their presence in structures, particularly bridges, is crucial, especially for state Department of Transportation (DOT) agencies overseeing various potential bat habitats. The project's core objective is to develop and validate a non-invasive method for accurately surveying bat species' presence in structures using DNA metabarcoding of fecal samples. The study will integrate passive acoustic monitoring, direct bat capture, and genetic techniques to enhance precision, offering an immersive research experience in bat conservation.
Network Mentors: Mandy Guinn, United Tribes Technical College
The Hunt for Insect Pollinators
Insect pollinators perform a key role in maintaining the health of agricultural crops and wild plants. More than 200,000 plant species rely upon insect-mediated pollination. Recent data have indicated that managed and wild bee populations and biodiversity may be on the decline. Although the critical role of bees and butterflies in pollination is widely recognized amongst scientists and non-scientists alike, very few are aware that a wide variety of insects from many orders and families are involved in pollination. However, the US lacks a coordinated nationwide surveillance network to collect critical data on our ~4000 native bee species as well as other pollinators. Moreover, no one has ever conducted a pollinator species survey in the Turtle Mountain region of North Dakota. For that reason, we started a pollinator survey of the Turtle Mountain region of North Dakota in the summer of 2022 and will continue it for at least the next four years.
Skills learned: plant and insect identification, insect collection, insect dissection, DNA extraction, PCR, DNA sequencing
Network Mentor: Scott Hanson, Turtle Mountain Community College
Some like it hot: do birds with warmer winter boxes nest earlier and produce more offspring?

In response to rising temperatures, many organisms have started breeding earlier in the spring. There is also evidence that individuals that experience milder over winter conditions breed earlier and have higher reproductive success. Yet, how variation in macro and microclimate conditions during the non-breeding season influence the timing of breeding and breeding success are not well understood and this information is becoming increasingly urgent in the face of climate change. This study will examine how variation in the macro (ambient) and nest microclimate temperatures influence the timing of breeding, parental behavior, and breeding success of free-living House Sparrows in Fargo, ND. This project will provide mentoring and training in the scientific process including hypothesis development, experimental design, data collection and analysis, and scientific writing as well as the opportunity to learn several skills including collecting, and analyzing nest temperature data, measuring parental behavior, and measuring nestlings in the field and collecting blood samples.
Network Mentors: Britt Heidinger, NDSU, Tim Greives, NDSU, and Sam Lane
Sharing the load:
how does nest temperature influence incubation behavior in male and female house sparrows?

Global ambient temperatures and the variability of temperature experienced by wild populations are expected to increase in the next century. This can dramatically affect oviparous animals, as eggs are more vulnerable to temperature changes, and require a narrow temperature range for successful development. Incubation behaviors can mitigate these effects; however, incubation is time consuming and energetically costly. Some songbirds provide biparental incubation, which can buffer their offspring from greater and more variable temperatures, while also decreasing the burden of care across pairs. However, we do not understand the ecological conditions or phylogenetic relationships that elicit maternal, paternal, and biparental incubation, nor how these life history strategies will change in response to climate change. This project will examine how experimentally elevated nest temperatures alter the bi-parental incubation behavior of house sparrows, and how maternal and paternal roles change in response to altered temperatures. Additionally, we will examine the patterns of incubation behavior strategies across the Americas, their phylogenetic relationships, and what ecological variables correlate with these patterns.This project will provide mentoring and training in the scientific process including hypothesis development, experimental design, data collection and analysis, and scientific writing as well as the opportunity to learn several skills including phylogenetic analysis, large data set curation, statistical analysis using R statistical software, collecting, and analyzing nest temperature data via radio frequency identification and temperature probes, and measuring adult and nestlings songbirds in the field.
Network Mentors: Britt Heidinger, NDSU and Samuel Lane, NDSU
Wildlife (Bird) Damage Management in Agroecosystems
A decline in songbirds from the 1970s until today has been documented with blackbirds (Icteridae) among the groups that have witnessed steep declines. While most of the US breeding populations have declined, blackbirds in the northern Great Plains have remained stable and are capable of forming fall roost aggregations of >1 million birds. In this region, blackbirds are considered an agricultural pest, causing >$3.5 million of damage to sunflower and $1.3 million of damage to corn annually as they feed on crops prior to migrating south. The mixed species flocks mainly include red-winged blackbirds (Agelaius phoeniceus), but also common grackles (Quiscalus quiscula), yellow-headed blackbirds (Xanthocephalus xanthocephalus), brown-headed cowbirds (Molothrus ater), and European starlings (Sturnus vulgaris). In collaboration with NDSU, the USDA-APHIS-WS National Wildlife Research Center focuses on studying nonlethal, socially-acceptable conservation actions to avoid native species from becoming pests while providing alternative habitat for foraging and spreading the birds (thus damage) across the landscape.
The following are 3 potential projects that a RaMP mentee could lead:
- Blackbirds are able to discriminate among sunflower achenes with differences in oil concentration as small as 5%, but removing hulls significantly impairs their ability to select achenes with higher oil content. The impact of anthocyanin, a chemical that creates an aversive flavor in achene hulls, has also been evaluated as a method to reduce blackbird damage by creating aversive sunflower varieties. Although oil content is likely more important for forage selection by blackbirds. Anecdotal evidence suggests that blackbirds may respond to the fatty acid composition (i.e., % oleic and linoleic) or the amount of unsaturated fat in sunflower varieties. Linoleic sunflower oil is a polyunsaturated oil, whereas oleic sunflower oil is a monosaturated oil. You will evaluate the foraging preferences of birds on sunflower achenes that vary in the percent oleic acid. Skills development: You will gain field experience in bird ID (by sight and sound) and monitoring of foraging preference of free-ranging wild birds or captive blackbirds. You will design and implement field research or aviary research evaluating foraging preferences of birds and gain experience analyzing data (R statistical language, composition analyses of oilseeds) and reporting results through oral presentations and written manuscripts.
- A reliable way to monitor blackbird distribution and abundance across broad scales during fall migration is needed. Effective monitoring of bird populations during the fall damage season will allow management tools to be tested with bird numbers as the response variable. Using bioacoustic monitors and game cameras, you will identify the presence/absence or abundance (sound amplitude, dB) of blackbirds in cattail roosts. Skills development: You will gain field experience in bird ID (by sight and sound), bird monitoring at roosts, and relationship of blackbird roosts with agroecosystem habitat. You will design and implement field research evaluating seasonal timing and behavior of blackbird roost aggregations and gain experience analyzing data (R statistical language, GIS, monitoring by soundscape and game cameras) and reporting results through oral presentations and written manuscripts.
- Blackbird damage is not evenly distributed within sunflower fields. Thus, an understanding of the spatial distribution crop damage is needed to efficiently mitigate the conflict. Taking a multiscale approach (i.e., crop, field, landscape), we can identify how bird damage varies spatially within a field and focus tools in high damage areas or bird entry points. Skills development: You will gain field experience in bird damage estimation in sunflower and its relationship to agroecosystem habitat at multiple scales. You will design and implement field research evaluating the best methodology for accurate damage estimation (infield estimates and tractor yield monitors); and gain experience analyzing data (R statistical language, GIS) and reporting results through oral presentations and written manuscripts.
This is a unique opportunity for individuals interested in the interface of agricultural systems and wildlife management. The position also provides applied experience for a career trajectory in local, state, or federal agencies or a research career focusing on the ever-growing field of human-wildlife conflict.
Network Mentor: Page Klug, USDA-APHIS-Wildlife Services
Ecosystem Homogenization of Prairie-Pothole Wetlands:
Understanding mechanisms and consequences of decreased biotic variability.
The Prairie Pothole Region spans across much of the northern great plains and gets its name from the millions of grassland embedded depressional wetlands (aka prairie potholes) scattered across the region. The wetlands and surrounding grasslands provide breeding habitat for over 50% of the North American duck population. However, wetland drainage, grassland loss, and climatic shifts have altered the ecohydrological variability of these wetland habitats. These changes have also led to changes in biological assemblages and likely facilitated biological invasions by fish and plants. Research is needed to understand the mechanisms that make these ecosystems vulnerable to climate and land-use change. Using long-term environmental monitoring data from the Cottonwood Lake Study Area, other U.S. Geological Survey studies, and additional data collection efforts, the trainee will be encouraged to explore research questions related to biological responses to ecohydrological regime shifts, landcover change, and identifying wetland types that are particularly important for conserving biodiversity. Depending on research interests the trainee will have the opportunity to work with field crews on several Northern Prairie Wildlife Research Center projects focused on wetland ecology. Specific projects the trainee would be expected to participate on include (a) collaborative project with Duck’s Unlimited investigation how conservation programs that utilize cover crops influence nutrient levels, aquatic-invertebrate communities, and breeding duck use and (b) a long-term wetland biomonitoring study investigating biological (amphibian, invertebrate, plant, and avian) responses to ecohydrological change.
Experimental tools and skills development: Experience with field-based environmental data collection in wetland ecosystem. Laboratory based processing of aquatic-invertebrate and water samples. Accessing and utilizing long-term datasets for analyses. Experience using R to analyze data. Summarizing and communicating research. Depending on the research questions, there is also opportunity for field or laboratory-based experiments.
Network Mentors: Kyle McLean, USGS
The Mile High Microbes
Airborne microbes have the greatest potential of spread among any organisms in most ecosystems. These microbes have unique characteristics and lifecycles which enable them to seamlessly survive in high altitudes or high atmospheric environments. These unique characteristics them the ability to withstand varying wind velocities, air pressures, air current speeds, variable and drastic temperature changes, and wet and dry conditions
The study of airborne microbes has renewed interest because of public health implications these microbes have as pathogenic organisms. COVID-19, Influenza, and many pandemic and epidemic pathogens affecting our societies today are spread primarily through airborne mechanisms. There is a huge concern that the ideal spread of bioweapons such as Anthrax will be effective if they spread through airborne mechanisms
Airborne microbes form part of the particulate matter fraction of the air, especially at the coarse (PM10) and fine (PM2.5) particulate matter fractions of the air. Their significance as part of any air quality assessment cannot be overstated. The current knowledge about microbial communities associated with airborne particulate matter, is limited. This study will fill this gap by describing the microbial communities associated airborne particulate matter using genetic sequencing techniques in combination with atmospheric science data culled from NASA’s TEMPO mission monitoring station at Sitting Bull College. Particulate matter will be sampled over 4 seasons in urban and rural settings. Then eDNA techniques will be used to extract and identify the microbes in the stratified air column of these settings. The project will have two main objectives and approaches: First, we will develop a standard operating procedure (SOP) for sampling airborne microbial communities in the atmosphere. The use of UVA technology and drones equipped with filteration and collection systems will be the focus pf this approach.Second, we will utilize eDNA techniques supplemented with next generation RNA-sequencing (RNA-Seq) to analyze the microbial communities.
Skills Developed: Students will gain hands-on field experiences with airborne particulate matter sampling, and use new technology in field research. Students would also gain valuable laboratory experiences in grnomis by working on eDNA extraction techniques.
Network Mentors: Mafany Ndiva Mongoh, Sitting Bull College
Genome-Wide Response to Environmental Stresses in Common Bean
Common bean is the most important food legume in the world produced for human consumption. It is rich in protein, fiber, and many macro- and micronutrients. The production of this important food source is challenged by a climate change conditions that are unpredictable. It can face flooding during early season germination, heat stress during the important flower portion of the growing season, and drought during the critical seed filling period of development. Detailed analyses of gene expression analyses are lacking for genotypes grow under these conditions. Better understanding of genome-wide expression pattern will provide plant breeders with selection targets to develop varieties better able to produce a crop under stressful conditions.
The goal of this project is to characterize the whole genome pattern of expression under abiotic stress (e.g. flooding and drought conditions). Controlled greenhouse experiments will simulate these various growth conditions. Additional field validations can follow. An RNA-seq expression study will identify suites of genes associated with various physiological pathways that are differentiate genotypes that can withstand or suffer from the stresses. The results of the experiments will identify allelic variants of important genes required for common bean to successfully survive the stress and possibly produce a crop under the stress conditions.
Professional development: During this project, you will learn: hypothesis development, project planning, and experimental time management skills.
Technical skills will include: greenhouse growth management, exposure to field-based research, tissue sampling, RNA extraction, RNA library preparation, and whole genome expression analysis using bioinformatics tools.
Personal skills will include: interacting with young and experienced researchers, verbal and written communicating scientific results, and public presentation of scientific results.
Network Mentors: Juan Osorno, NDSU and Phil McClean, NDSU
Morphological and Physiological Response of Barred Tiger Salamanders to Fish Introductions

Barred Tiger salamanders (Ambystoma mavortium) are a polyphenic species. Aquatic larvae respond to variable environmental conditions by metamorphosing into terrestrial metamorphic adults that will leave their natal waterbodies or larvae will retain their larval morphology and become sexually mature paedomorphic adults that maintain their aquatic adaptations (i.e., gills and finlike tail). In some case, certain environmental conditions, such as increased competition for invertebrate prey has been linked to a unique “cannibal” morphotype. Cannibal morph larvae can be distinguished from typical larvae by their large spade-shaped heads and the presence of enlarged vomerine teeth. In most cases cannibal morphs have been found in drying ponds with limited prey resources and an overabundance of tiger salamander larvae. The large heads and vomerine teeth enable larvae to eat larger prey in these environments, such as other salamander larvae, hence the name “cannibal morph”. In North Dakota, cannibal morphs have been observed in large wetlands, typically containing an abundance of small fish such as fathead minnows (Pimephales promelas) and low abundances of salamander larvae. The resulting hypothesis is that resource competition with small fish can also induce cannibal morphology. We propose to have participants carry out a experiment using mesocosms or aquaria to attempt to test this hypothesis. The experiment would involve collecting eggs or very young salamander larvae and rearing them with fathead minnows and measuring physiological stress and morphological expression over time. The participant will also be able to utilize previously collected tiger salamander, invertebrate, and hydrological data to identify if the presence of cannibal morphs is correlated to fish and salamander abundances, resource availability (invertebrates), hydrologic conditions.
Experimental tools and skills development: Field methods for amphibian work and analysis of physiological characteristics such as hormone levels (glucocorticoids, thyroid hormones) at critical life-history stages. Experimental design using aquatic mesocosms. Data extraction from a large long-term database and analysis tools for analyzing complex and large datasets with environmental variables.
Network Mentors: Travis Seaborn, NDSU and Kyle McLean, USGScollaborating with Tim Greives, NDSU
How the elk herd roams:
investigating population structure and connectivity using genetics
A central question in biology is understanding how landscape and geography influence the connectedness of populations. Although radio telemetry methods can help us understand movement patterns, it does not always directly relate to dispersal events that lead to mating and, consequently, gene flow. This is particularly true for large mammals. In some large mammal systems, individuals may move long distances but do not necessarily breed with individuals outside of their local population. One way to estimate true gene flow and understand how connected populations may be is using genetics. In northern Minnesota, northeastern North Dakota, and southern Manitoba, the amount of gene flow and population structure among elk hers is currently unknown. The connectivity of populations and/or subpopulations have important management implications for understanding population and movement dynamics. For these particular herds, they currently are managed under different strategies (e.g. no hunting of the Manitoba herd), so this research will have important management implications.
To explore these ideas, the student will have the opportunity to use genotype-by-sequencing data from more than 500 elk samples coming from across areas in Minnesota, Manitoba, and North Dakota. The student will work with mentors to estimate population genetic differentiation and gene flow, with a potential emphasis on how these characteristics relate to geographic distance. The student may also explore metrics of ‘genetic health’, including things like inbreeding coefficients, effective population size, and heterozygosity.
Experimental tools and skills development: Genomic data filtering and data wrangling, geographic information systems (GIS) modeling, population and conservation genetic analysis.
Potential Network Mentors/Collaborators: Travis Seaborn, NDSU; Levi Newediuk, University of Manitoba; and Michelle Carstensen, Minnesota Department of Natural Resources
Small RNA, Big effects!
Changing environments means species are moving around more than in the past - both because of humans moving around and the climate changing. When species move around more, they meet new species they haven’t met before. When that happens, they get new diseases. Not just bacteria and viruses, but also bits of DNA that live inside their cells and replicate selfishly.
Small RNA is how organisms defend against these invasions. We will work on how small RNA populations evolve in response to new invaders.
Skills that will be acquired during the RAMP:
Molecular biology: PCR, DNA/RNA extraction
Bioinformatics: basic coding skills
Technology: Operating next gen sequencing technology
Network Mentors: Sarah Signor, NDSU


